
Chapter 5 Phylogenetics basics
5.1 How to read a phylogenetic tree
A phylogeny, or phylogenetic tree, is a diagram that shows the evolutionary history and relationships among or within groups of organisms. Phylogenetics was traditionally a somewhat obscure field in which systematists (biologists concerned with arranging organisms into a tree that showed their ancestral relatedness) arranged related living organisms at the tips (or “leaves” of the tree), and made branches to connect different organisms back to putative ancestral organisms.
Here’s a phylogeny of the family Ursidae (the bears).
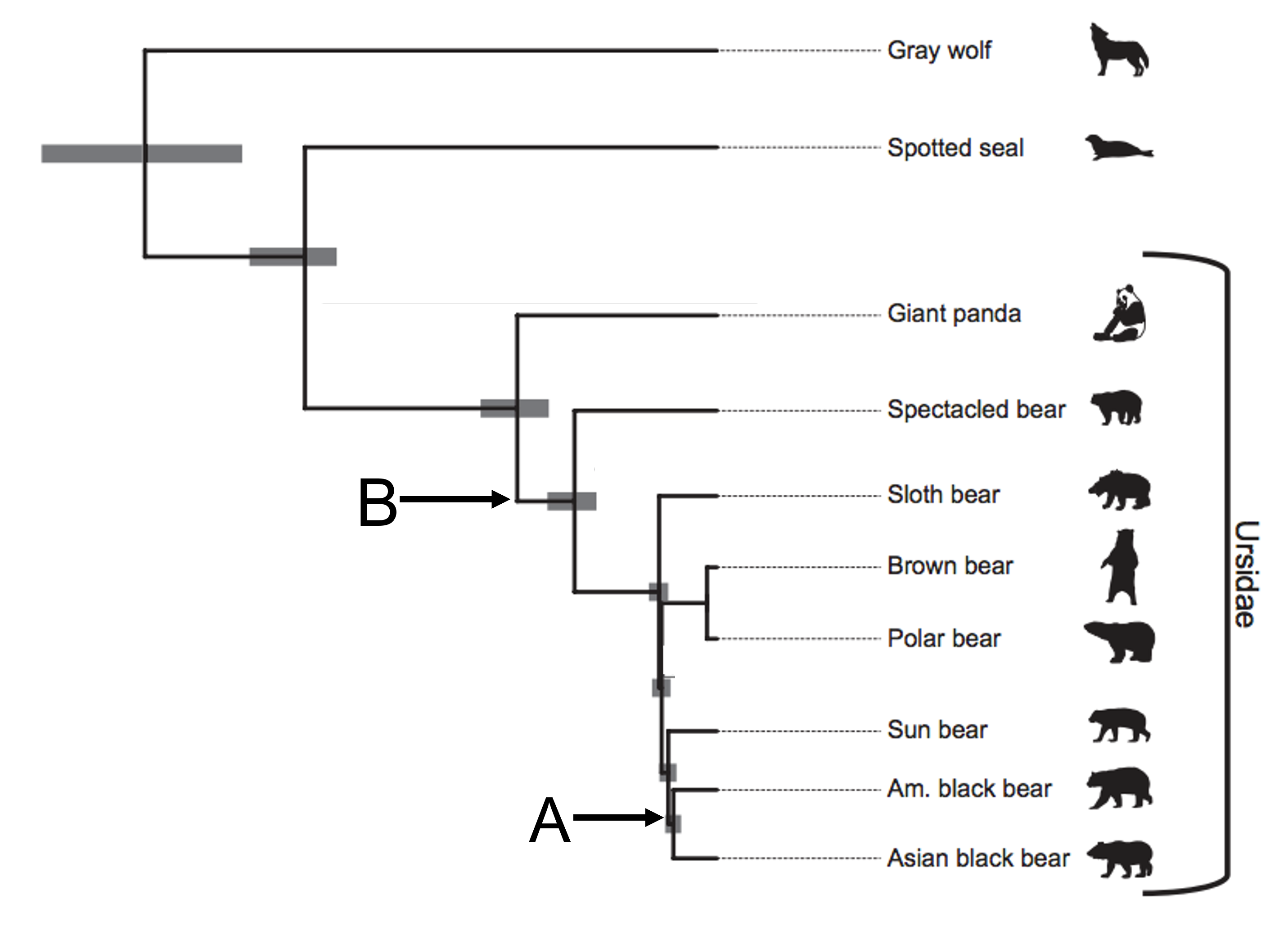
In this tree, all the extant species (or currently living species) are at the tips on the far right side of the phylogeny. Inferences about how the bear species are related become apparent as you move away from the tips down the branches. When two branches meet at a node (as they do at point A), you can assume the species at the tips of those branches share a common ancestor. For example, this phylogeny of the Ursidae indicates that American black bears and Asian black bears share a common ancestor (indicated by the node at point A). However, we don’t know what the common ancestor is for certain, we are just inferring based on similarities between the species that exist today.
Nodes that are closer to the tips indicate species that are more closely related (and thus indicate a more recent common ancestor than nodes farther away from the tips). American black bears are more closely related to Asian black bears than to American black bears are to giant pandas, because the American black bear branch connects to a node shared by the Asian black bear branch (point A) before it connects to a node shared with the giant panda branch (point B).
Another unusual thing about phylogenies is we can change the order of the taxa on the tips without actually changing the topology of the tree.
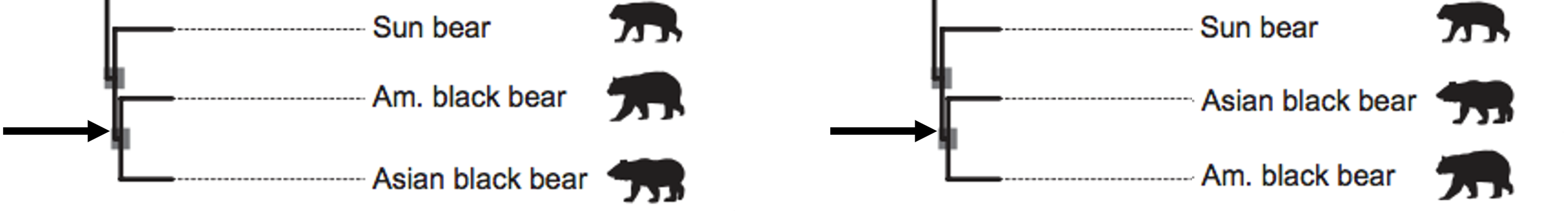
These two trees are the same, even though we have changed the position of the labels of American black bear and Asian black bear. In phylogenetic trees, relatedness is expressed by the distance to a common node between two species, NOT by whether the labels are near each other. Branches can rotate freely around nodes without changing the tree.
5.2 Outgroups
Although this is a phylogeny of the Ursidae, you might have noticed there are two branches belonging to the gray wolf and the spotted seal, neither of which is a bear. These two species are included as outgroups. Outgroups are taxa that are only distantly related to the group of interest and serve as reference points for determining evolutionary changes.
5.3 Branch lengths
Branch lengths (the distance between two nodes, or between a node and a tip), may or may not be indicative of the passage of a particular amount of time. It depends on how the tree was inferred (we infer phylogenetic trees, we don’t make them). If the tree is created by parsimony or neighbor-joining methods, the branches simply indicate that there was one (or more) change from the ancestor to the descendant. If the tree was created using maximum likelihood methods, the branch lengths represent how many genetic changes occurred over time.
Regardless of how the trees are constructed, they are estimates of what we think happened historically. Each estimate contains within it implicit assumptions about rates of mutation accumulating, likelihood of different types of changes being more common (transitions vs. transversions, for example), and so on. The tree is our best hypothesis as to the history of the organisms on it, but it is only a hypothesis.
At one time, only morphological data could be used to make these trees. Thus, phylogenetic trees might have been based on similarities of bone structures, or fur types, or other gross physiological features. Even though the trees were called “phylogenetic” trees, they were not based on genetic data.
Now, phylogenetic trees are generally based on DNA sequence (for closely related species) or amino acid sequences (for more distantly related species). Furthermore, the trees are generally based on several genetic loci, rather than on the whole genome. This is changing, with next generation sequencing and advances in computing power. Nevertheless, at present most phylogenetic trees are “gene trees” rather than “species trees,” and it is important to remember that selection or drift on a particular locus can influence a tree so that it reflects the history of the gene, but NOT the history of the species