
Chapter 8 Finding sequences in GenBank
8.1 Identifying a query sequence on GenBank
For this book, we will use Glu-1 sequences from a variety of species to infer our tree. Glu-1 is a gene that encodes one of the subunits used to make gluten in plants like wheat. We will use this gene to reconstruct some of the deeper phylogenetic relationships among the grasses.
We’re going to temporarily leave AnVIL and RStudio and head to NCBI’s website.
We start by searching for Glu-1 sequences in the NCBI nucleotide database. At the top of the website, use the pulldown menu to choose “Nucleotide” and enter “glu-1” in the search bar.
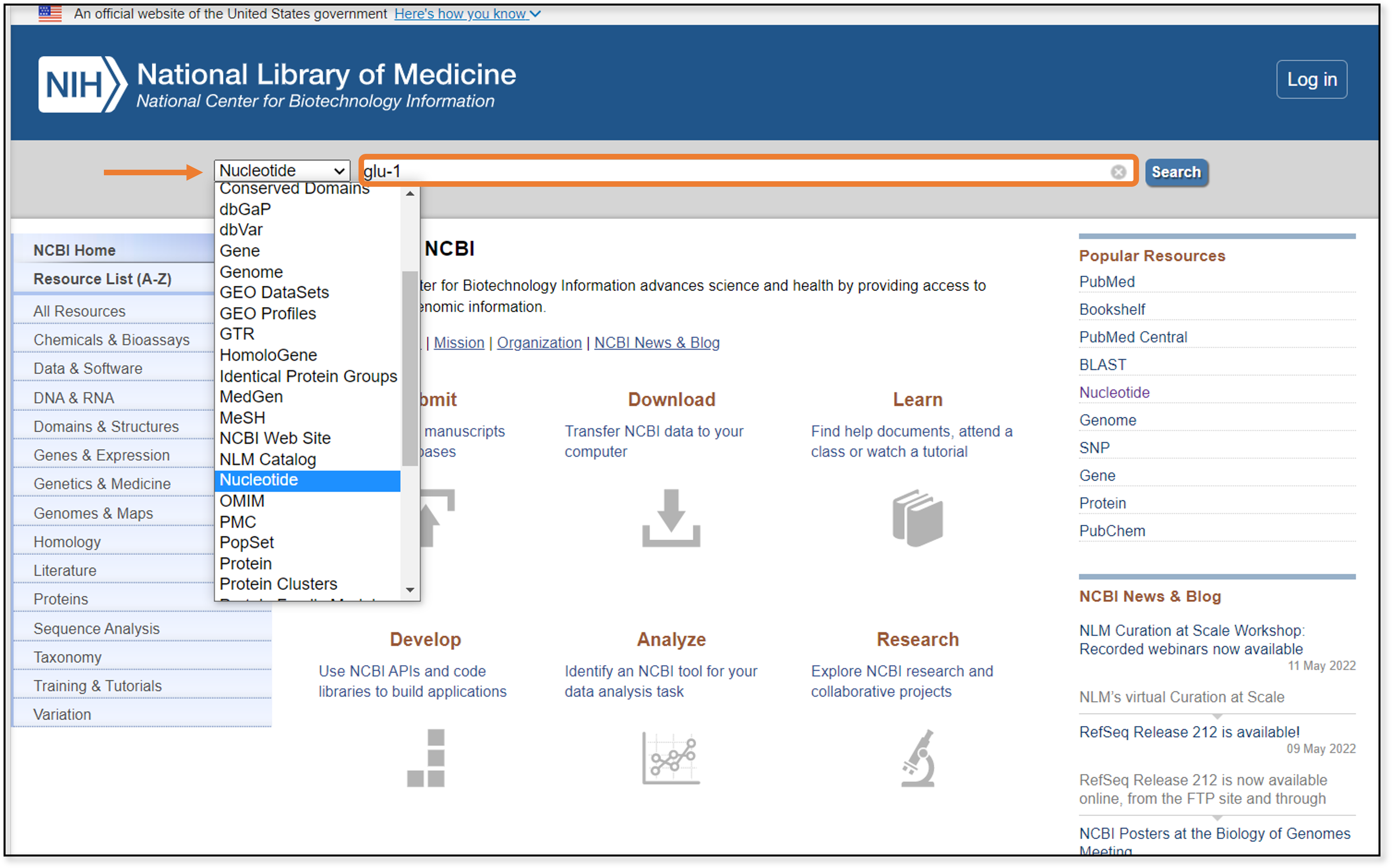
You might notice that this returns thousands upon thousands of possible sequences. While it’s nice having choices, having too many results makes it difficult to know where to start. Instead, we’re going to narrow down our sequence choices by specifying that we want Glu-1 sequences from common wheat, or Triticum aestivium. This is a good starting point, since we know that common wheat plants make the gluten protein, so the genome should contain Glu-1.
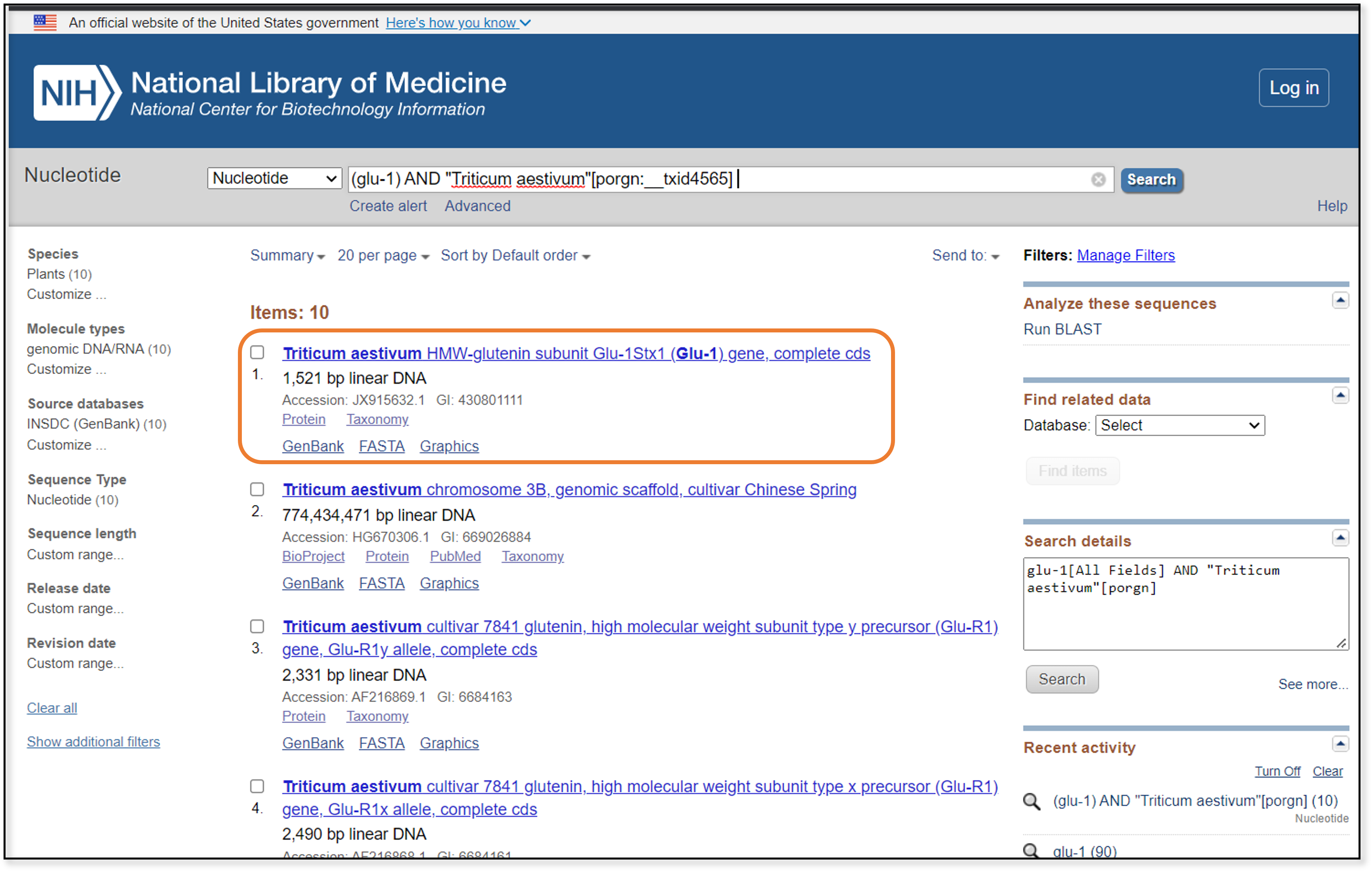
The first hit (at least from when this guide was created) is exactly what we’re looking for - the complete coding sequence for the high molecular weight glutenin subunit, the Glu-1 gene. If we click on the link at the top of the entry, we can go to the GenBank page for this particular entry.
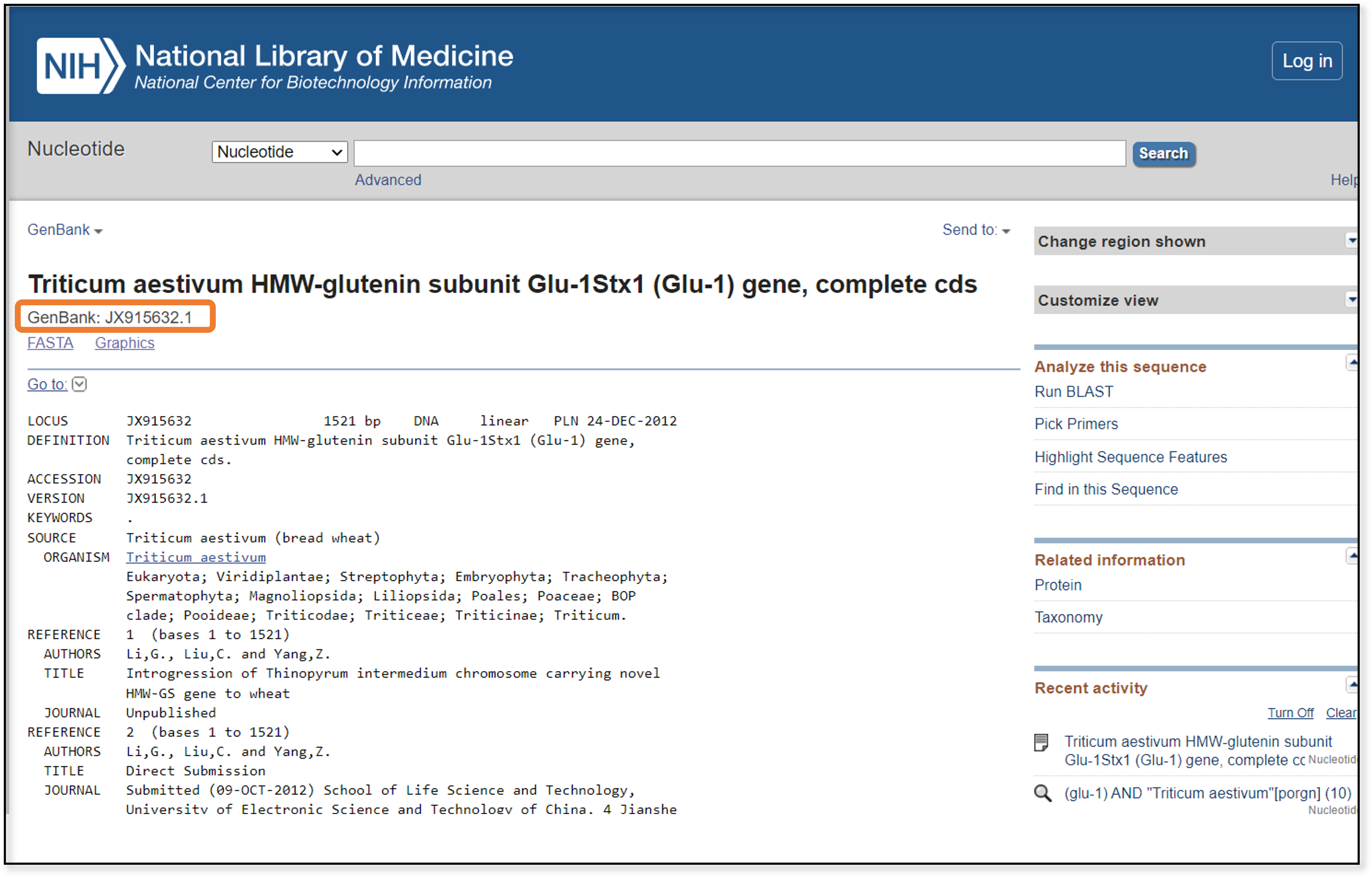
This page contains a lot of information about the sequence, including which research group generated it, if the sequence was used in published research, and the full taxonomy of the sample. At the top, we also find the accession number, or the unique ID assigned to this particular sequence. Highlight and copy the accession number - this is what we will use for our next step, a BLAST search.
8.2 blastn
NCBI created a tool that allows us to use the basic local alignment search tool (BLAST) algorithm to find sequences similar to our query sequence (in this case, the Triticum aestivium sequence we identified above). Here’s a link for NCBI’s web tool: BLAST.
There are many tutorials on how to use BLAST (including NCBI’s own), so this section is going to focus primarily on the logic behind choosing sequences for phylogenetic analysis, not just the steps.
Once you open the BLAST webpage, you have five options for searching (the tabs at the top of the page). Which method you choose depends on your query sequence. We’re going to work with two of them: blastn, which identifies DNA sequences that are most similar to the DNA (or nucleotide) query sequence; and blastp, which does the same for protein sequences.
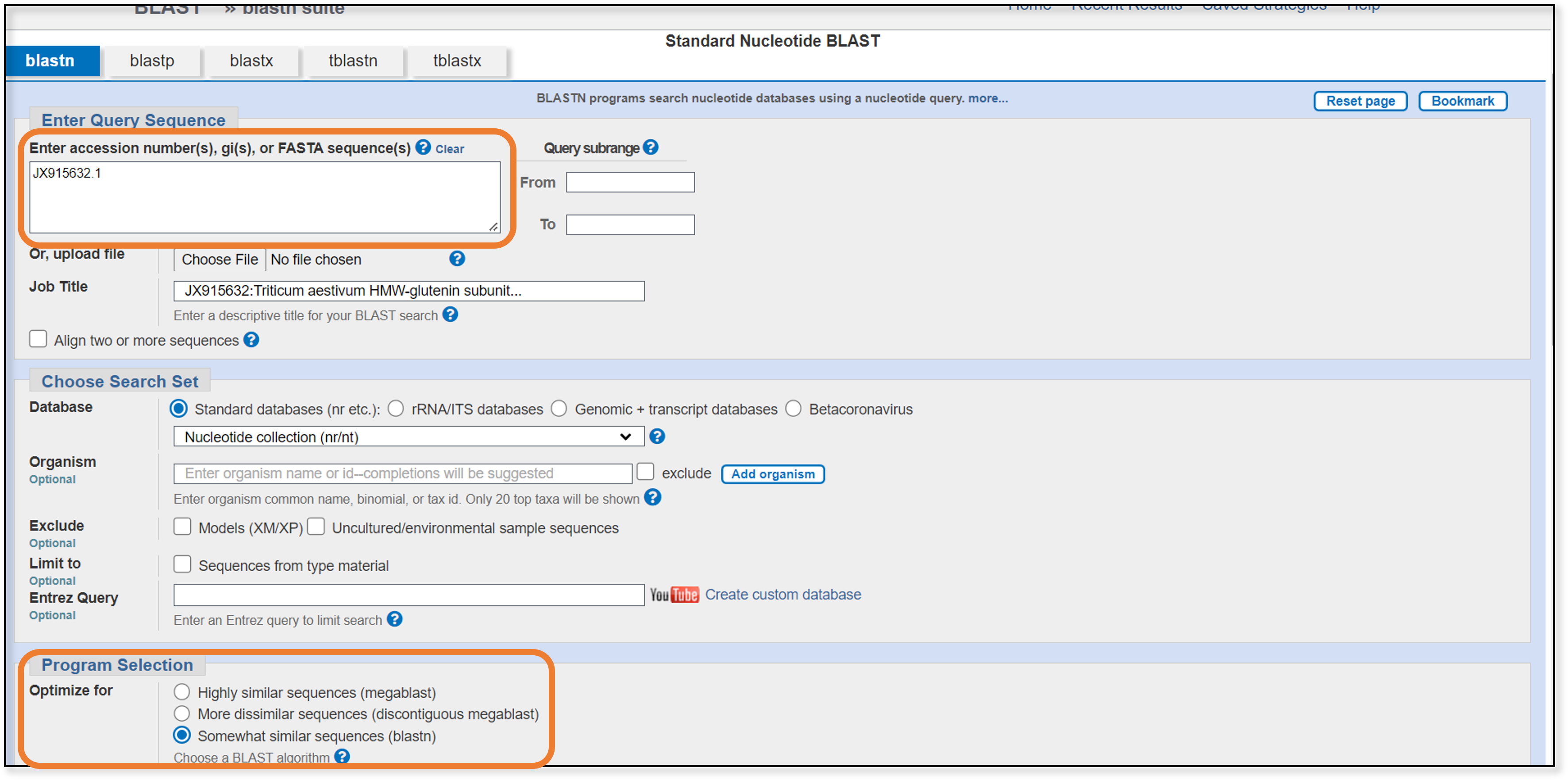
For the blastn search, all we need to do is paste the accession code from earlier into the search box and change our program selection to somewhat similar sequences (blastn). Next, let’s go down to the bottom of the page to the algorithm parameters section.
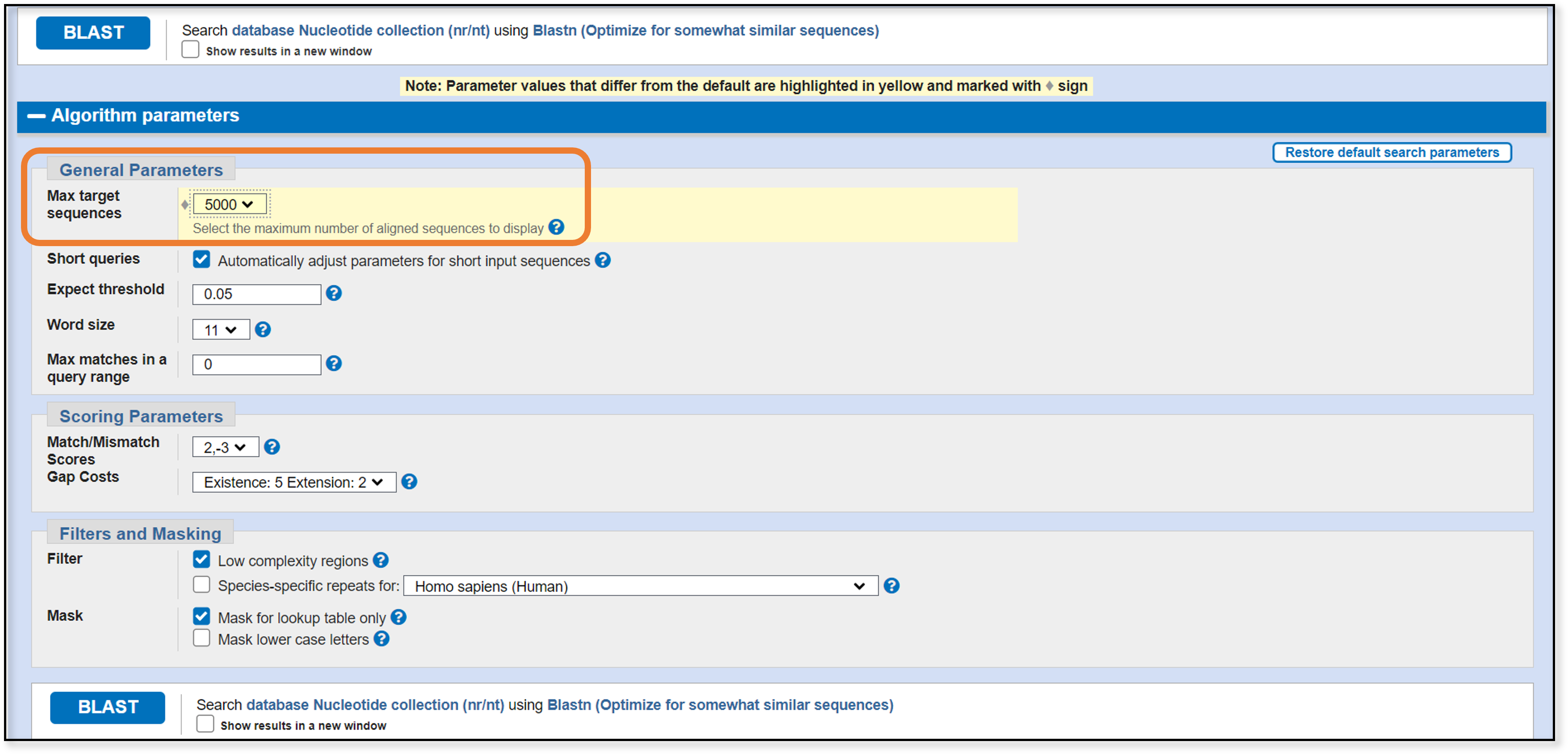
We need to change the max number of target sequences (the maximum number of sequences for our search to return). Given how rapidly the size of the GenBank databases are growing, leaving this value at 100 means we will miss a lot of sequences that we might otherwise want to see. For now, we can leave the other parameters as the default settings. The click the BLAST button on the bottom left.
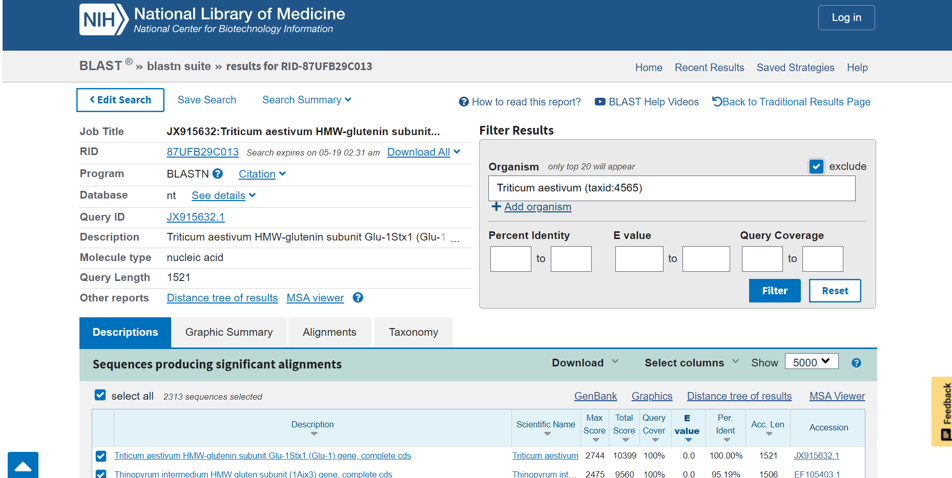
It can take a couple of minutes for the blastn search to finish. When it does, a webpage similar to the figure above will open. On the right side of the screen, we have the option of applying additional filters to our search. Because we are interested in looking at the deeper phylogenetic relationships among the grass family, we don’t necessarily want any additional Triticum aestivium sequences, so we will filter them out. That leaves us with over 2,000 other sequences from which to choose our taxa. (If you were interested in more shallow phylogenetic relationships, choosing multiple sequences from the same taxa, or dense taxon sampling, is a good decision.)
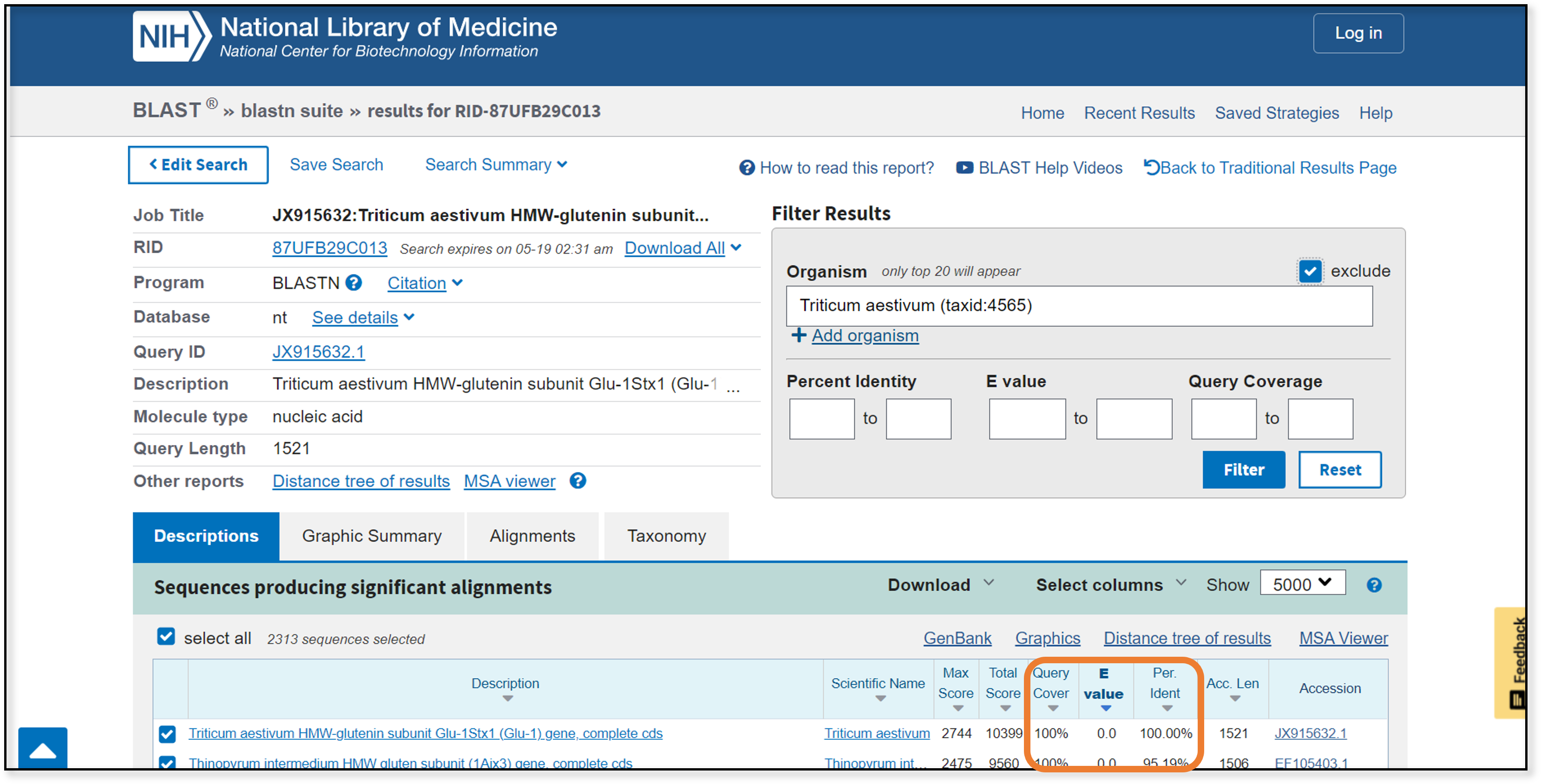
There are three quality-control statistics at which we want to look. query cover: the amount of overlap between our query sequence and the newly-aligned sequence; larger is better _E value_ (expect value): the number of hits expected by chance; like p-values, a lower number is better *per ident (percent identity): the percent similarity between the two sequences; larger is better
We can filter or sort on any of these statistics. At this point we need to really look at the aligned sequences and decide which ones we want to use.
There are quite a few samples from a variety of grass species that show good overlap, low E values, and high percent identities. Since we have options, we will prioritize choosing samples with complete coding sequence whenever possible (and avoid any sample labeled “pseudogene”, since that isn’t the actual Glu-1 gene sequence).
We will focus on these 9 sequences (in addition to the common wheat sequence we identified earlier):
- EF105403.1, Thinopyrum intermedium (intermediate wheatgrass)
- DQ073553.1, Leymus racemosus (mammoth wild rye)
- EF204545.1, Lophopyrum elongatum (tall wheatgrass)
- AJ314771.1, Secale cereale (rye)
- FJ481569.1, Henrardia (a genus of Asiatic wheatgrass)
- DQ073533.1, Agropyron cristatum (crested wheatgrass)
- AY804128.1, Aegilops tauschii (Tausch’s goatgrass)
- AY303125.2, Taeniathetum caput (medusahead rye)
- KF887414.1, Dasypyrum villosum (mosquito grass)
A quick check of the taxonomy confirms that all of these samples are from the grass family, family Poaceae.
8.3 Identifying an outgroup
We have two approaches we could take for identifying an outgroup - we could use a more distantly related taxon, or we could use a homologous gene sequence from a more closely-related taxon. When we look up information about the Poaceae, we find there are three clades within the family - cereal grasses (like wheat), bamboos, and grasses (such as those species found in natural grasslands or cultivated for lawns and pastures). In the list of 10 related sequences above, we don’t have any sequences from the bamboos (subfamily Bambusoideae). Glu-1 from a bamboo species might make a nice outgroup, if we can find a sequence for it.
First, we’ll try another blastn search, this time setting the program selection to more dissimilar sequences (discontiguous megablast)

When we get those results back, we can filter for samples within the subfamily Bambusoideae. Alas, we have no sequences that match.
The next thing we can try is a blastp search. These searches are nice for identifying more distantly related samples, because the protein sequence of a gene changes more slowly than the nucleotide sequence. In order to run a blastp search, we need a protein sequence for our query. Luckily, we chose a full coding sequence. When we look at the GenBank entry for JX915632, we can find the coding sequence translated into the amino acids at the bottom of the page.
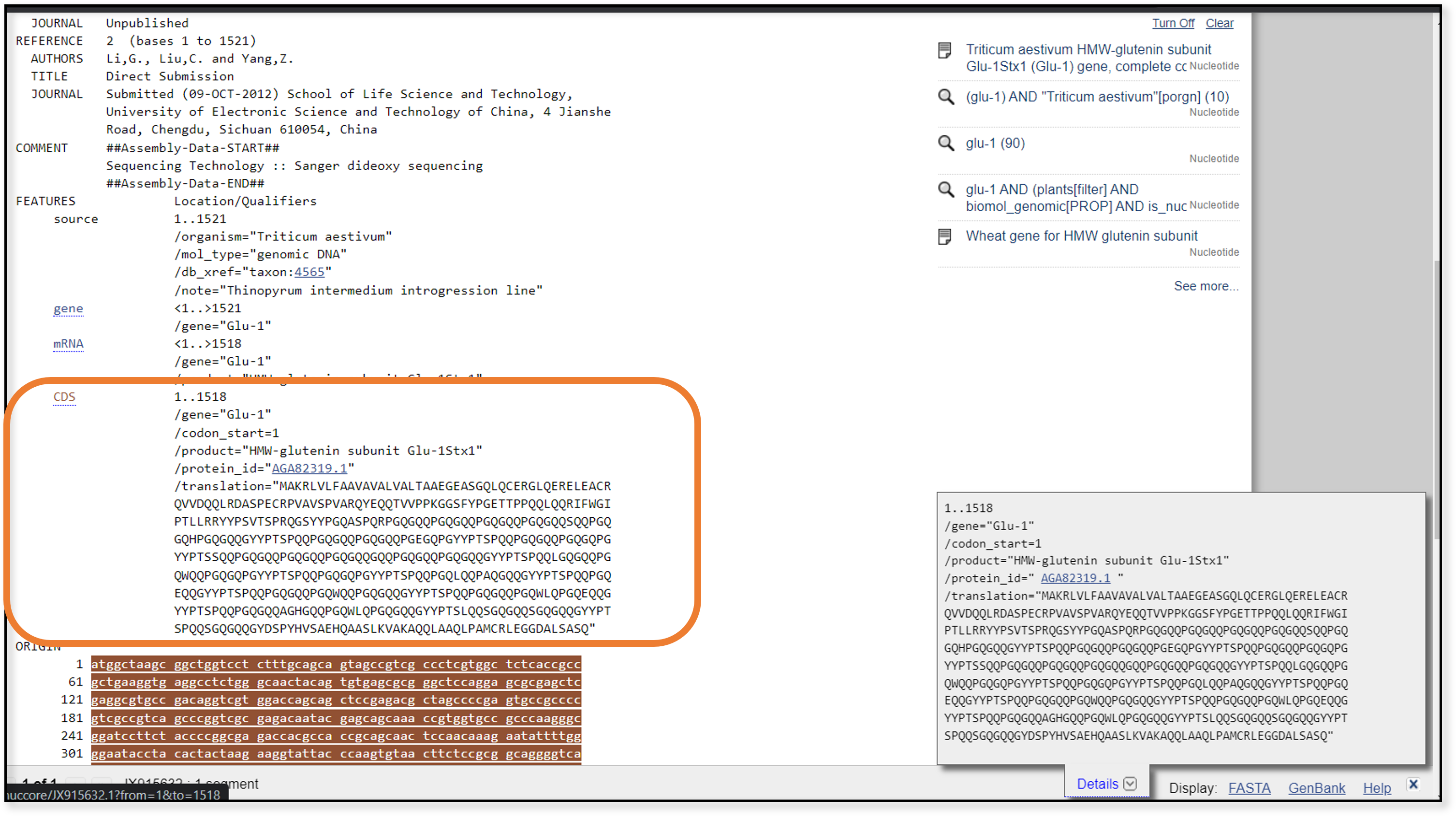
We can copy this amino acid sequence and paste it into the query box on the blastp page.
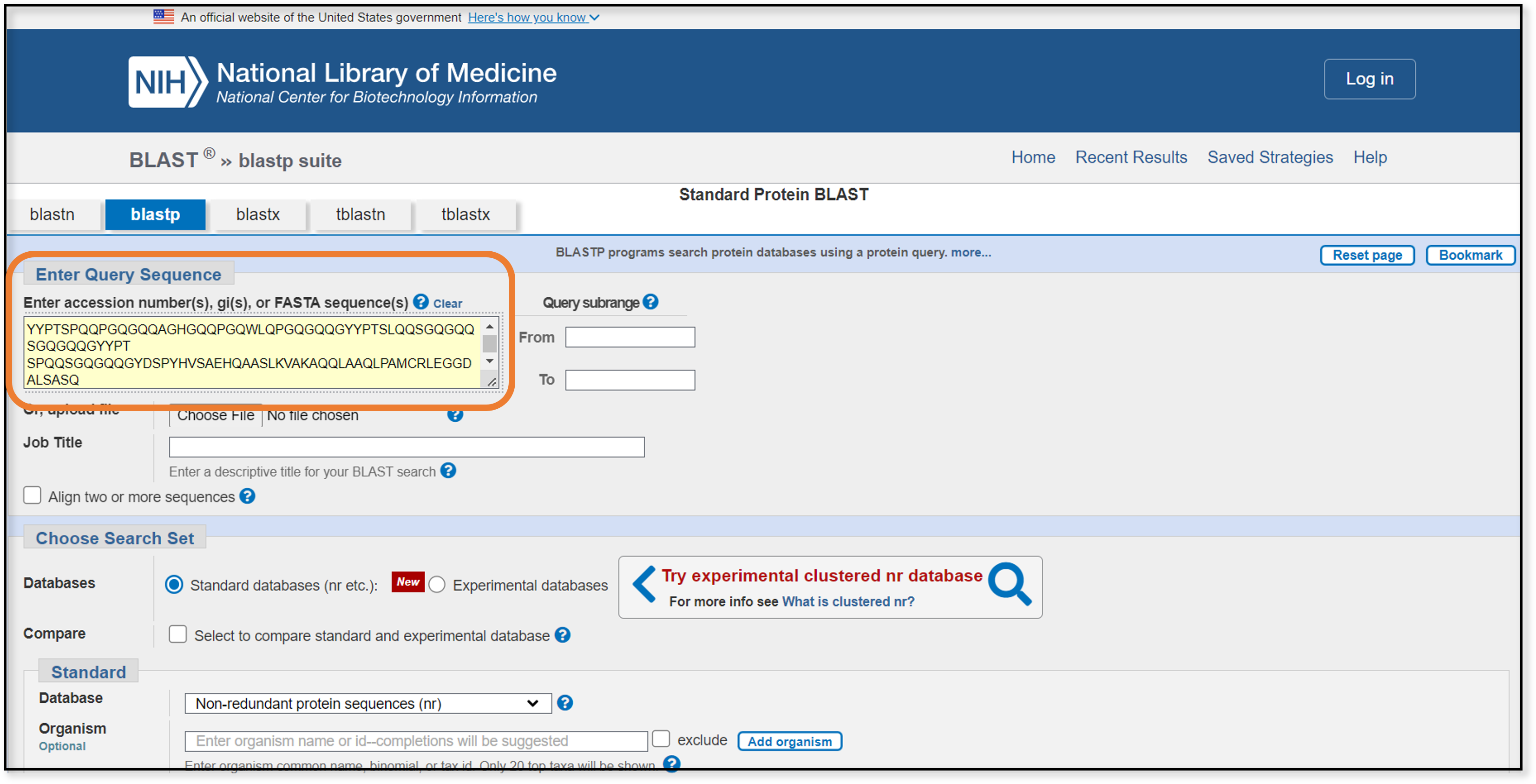
After the blastp search finishes and we filter out Triticum aestivium results, we end up with several hundred matches. Great! …or is it?
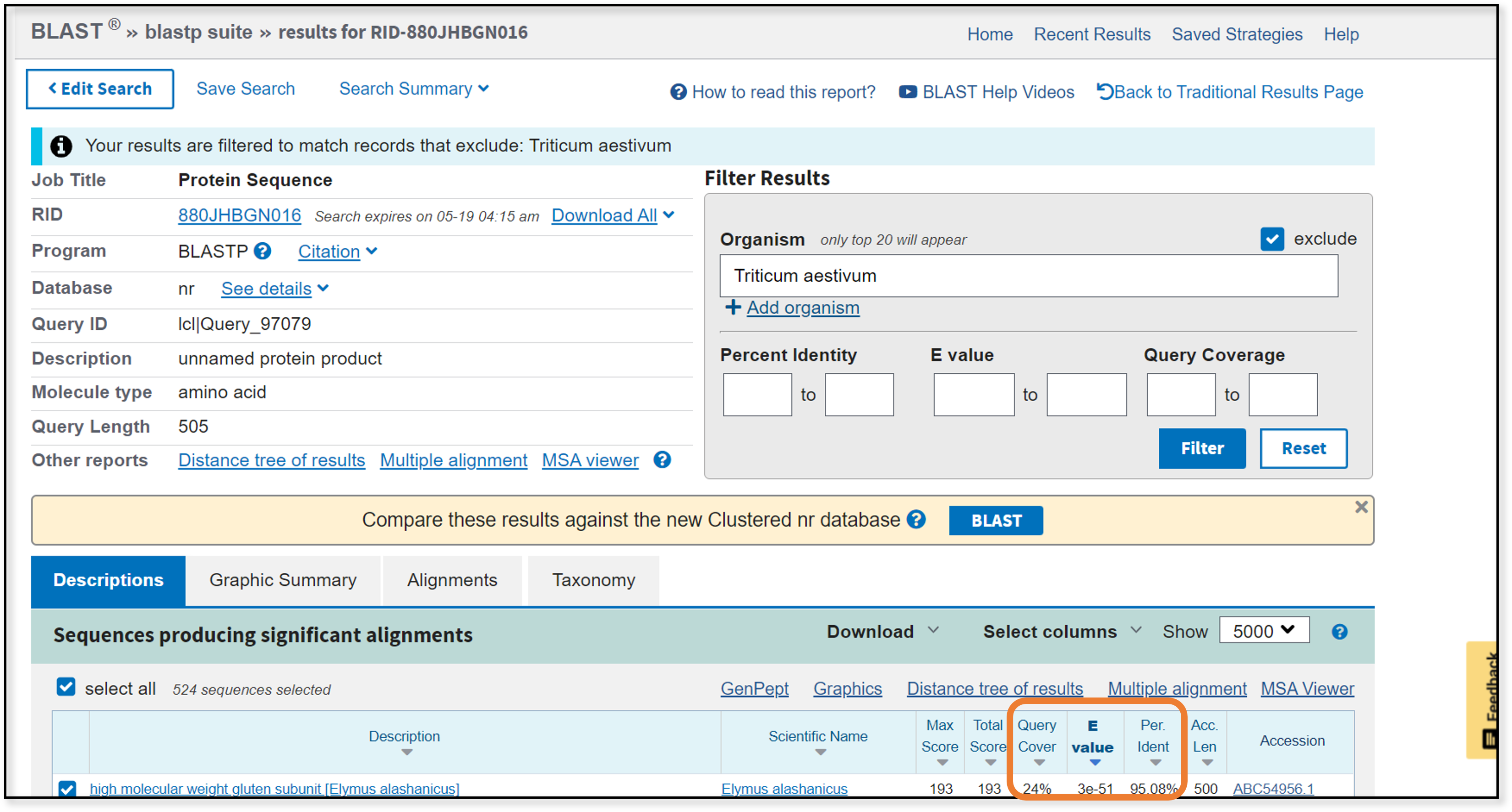
Unfortunately, all of the samples that are returned have very poor query coverage (less than 25%). None of these samples are likely to work for our purposes. Instead, we will have to try a homologous gene from a closely-related taxon. In our first blastn search, samples labeled “D-hordein” showed up near the bottom of the results. A Google search suggests that D-hordein is a barley homolog to the wheat Glu-1 gene product. This might serve nicely as an outgroup.
We will add 2 additional sequences to our list, for a total of 11:
- D82941.1, Hordeum vulgare (barley) D-hordein
- JX276655.1, Elymus sibiricus (Siberian wild rye) D-hordein