Chapter 7 Computing Resource Decisions
Now that we have discussed a bit about how computers perform computations and described a bit about computing options, let’s discuss more about how you might choose the right computing resources for your work. In this chapter, we will discuss aspects that you should consider when deciding between different computing resource options.

To help you make an informed decision about computing resources, it is useful to become familiar with the benefits and drawbacks of various computing options. First, we will start out with some general considerations that you should think about when beginning to determine what computing option makes sense for your work.
The following are the major decision points for your computing needs:
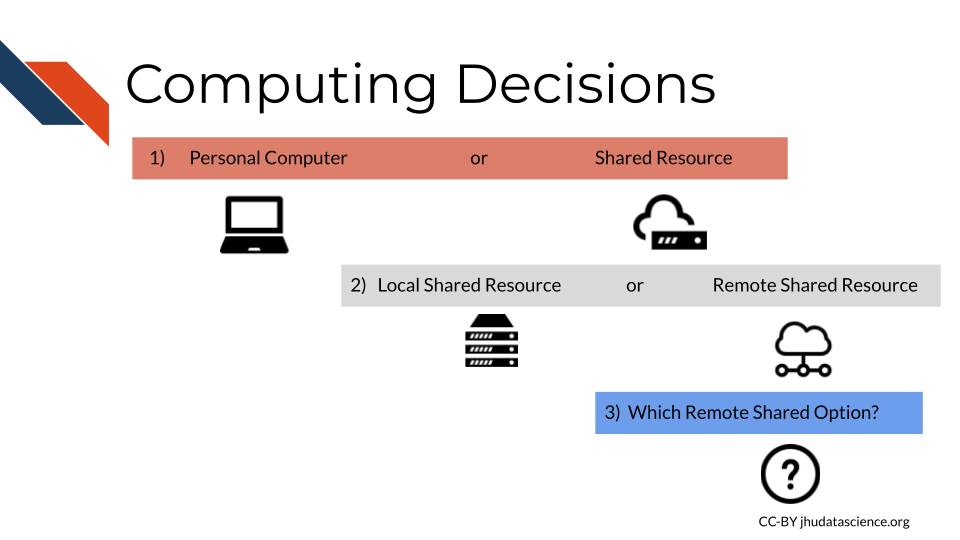
Note: This content was adapted from content by Frederick Tan for the AnVIL project. See his course created with Jeff Leek, Sarah Wheelan, and Kai Kammers here.
7.1 General Computing Considerations
Choosing a computing platform depends on several considerations.
Asking yourself and your research team about the following considerations can help you find the right computing platform.
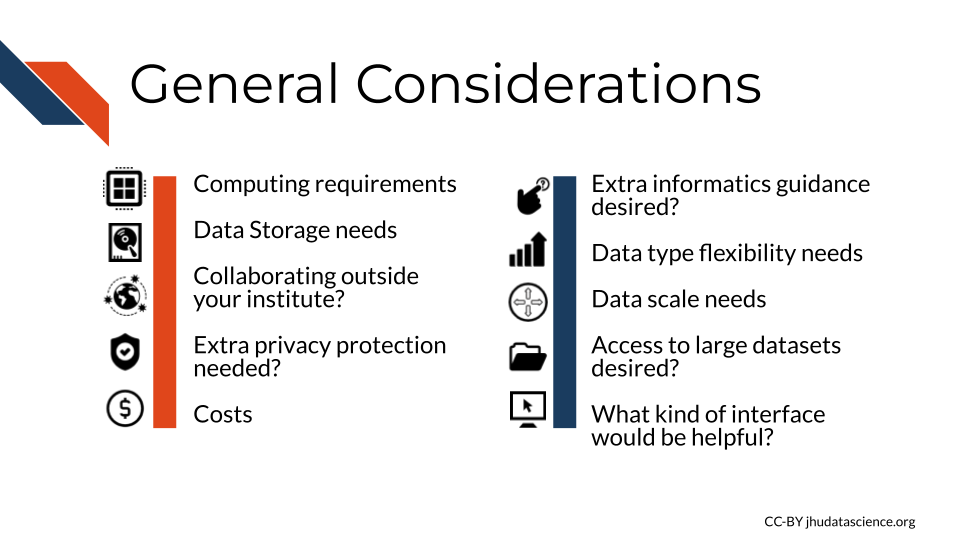
Let’s take a bit of a deeper dive now for each of these considerations.
7.1.1 Computation needs
Now that you know more about determining your personal computer’s computing and storage capacity, as well as how to determine or estimate the files sizes that you might use for your research, you can begin to assess if your personal computer is up to the task. When determining what your computing needs might be, remember to evaluate how many files you might use in your analyses, the file sizes, the amount of RAM and CPUs (and possibly GPUs that your computer has) and some level of understanding for how intensive the computing tasks are that you plan to perform. How do you assess this? If the files that you intend to use in your analysis are quite large for your computer’s storage capacity, then it is likely that your computer might struggle to work with such files. This might also be the case if you plan to use many smaller files (such as hundreds or thousands, but smaller files can add up quickly). Finally, if you plan to perform many steps on your files in your analysis, this may also require more computing resources than you have available on your current personal computer. Shared computing options will generally have the capacity to allow you to do your work, unless you have very large data needs. If you have plans to analyze large datasets, it would be a good idea to check the computing capacity of the local or remote computing options that you are interested in before you start an analysis. Cloud computing options can be great if you need more efficiency, as there are no job queues to worry about like with more traditional shared resources.
7.1.2 Data storage
Again, now that you know how to assess the data storage potential of your computer, you can decide if your computer can handle storing all the files that you might wish to use in your analysis. Think about your current data analysis plans, but keep in mind your future plans as well. If you hope to replicate experiments with more samples, you might run out of storage. One way around this is to by external additional storage (which is also a good idea for backing up your data!). However, if you think that you might have much larger scale research plans in the future, you might want to think about shared computing options. Cloud computing platforms and more traditional servers have different storage capacities, so it is worth checking out the options that might be helpful for your research. Also keep in mind that it will take time to transfer your data, especially if your data is very large.
7.1.3 Multi-institute collaboration
If you plan to work with others outside of your institute that would not have access to the same local shared computing resources, then remote computing options would be really helpful for allowing your collaborators to work on the same data together. Cloud platforms make it especially easier for collaboration, as everyone can share the exact same computational environment, including hardware, software, and datasets.

7.1.4 Protected data
Are you working with protected data that requires special security precautions?

[source]
If you are working with data that might be protected by HIPAA, such as electronic health records, then special security measures are required to ensure that only authorized users have access to the data. Be careful about both local and remote shared resources. Sometimes extra privacy and data security measures are built in and at other times, you will need perform extra steps to get the data security and privacy that you need. Although many cloud computing systems do not allow for extra privacy, there are platforms that provide compliance with regulations like HIPAA and FedRAMP.
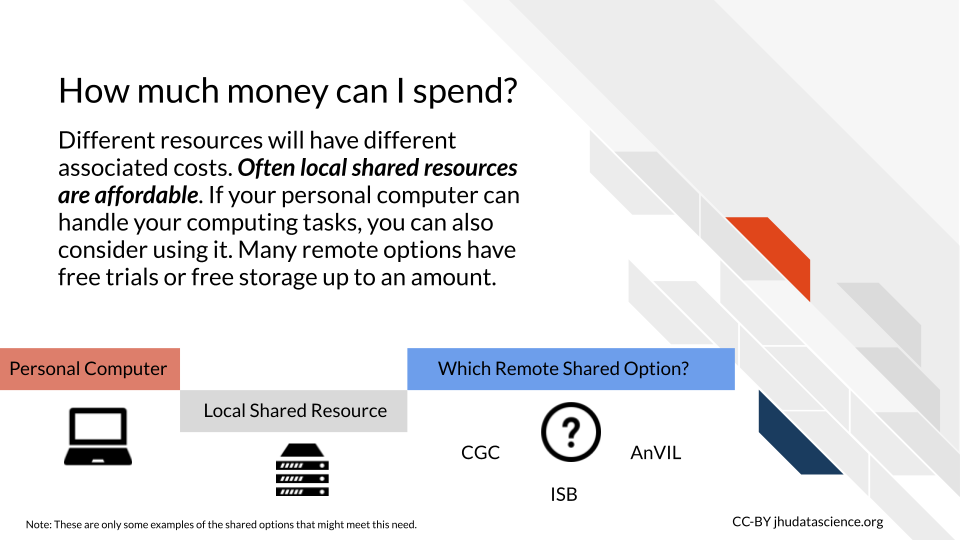
7.1.5 Costs
Often local shared computing resources at your institute can be much less expensive than some of the common cloud computing options. However, this is not always the case and if you have very specific analysis goals in mind, the benefit of cloud computing resources is that you typically only pay for the resources that you actually use. This also involves learning how costs are calculated for the particular cloud resource, which can be a challenge, but many cloud platforms that were designed for research such as Jetstream or Terra/AnVIL can be very affordable; also, some small platforms offer free resources or free trials initially to start.
7.1.6 Extra guidance
Cloud computing platforms such as Galaxy, AnVIL and GenePattern offer lots of training material and resources about how to actually perform analyses, especially genomic analyses. Galaxy also supports other types of data, as do many other platforms, as described in the last chapter. Having the extra guidance like that offered with these types of platforms can be very beneficial to investigators that are trying out new methods!

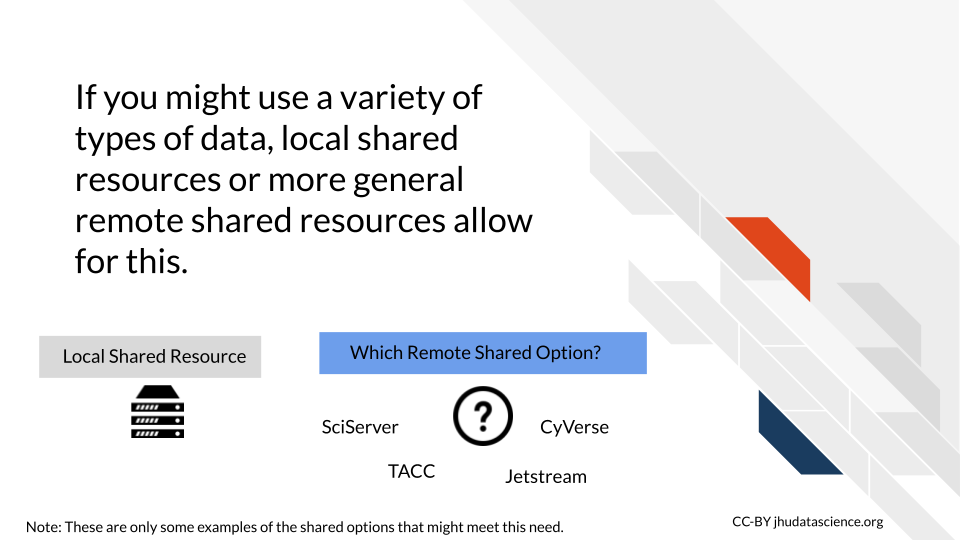
7.1.7 Flexibility
If you plan on working with multiple data modalities like for example both imaging and genomic data, consider computing options that are flexible for such analyses. Some cloud computing options designed for research are more general and supportive of this, such as Galaxy to some extent, as well as to a larger extent SciServer, Jetstream, or CyVerse.
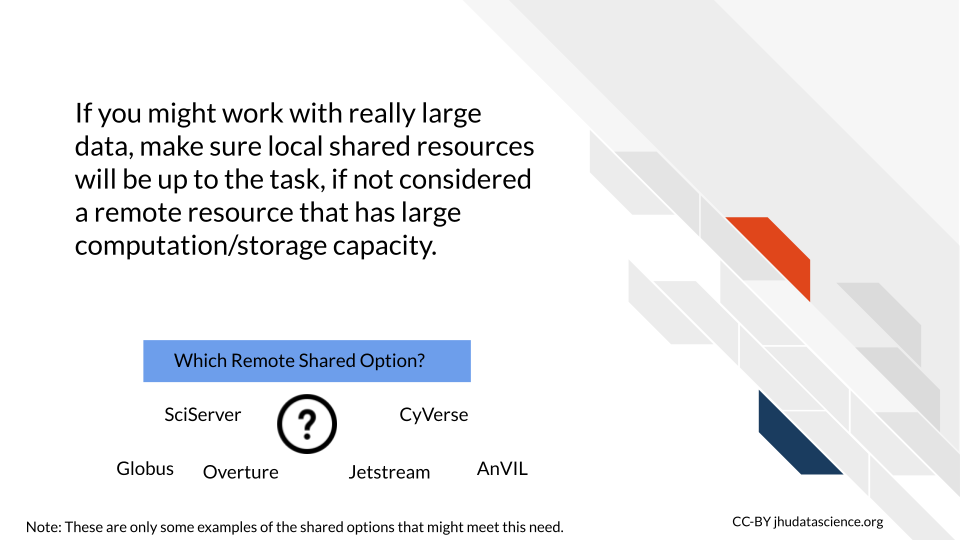
7.1.8 Scalability
If you hope to start using very large datasets or plan to collaborate with many people using many different data sets, then you might need to keep in mind the future storage and computing capacity of the computing resources that you are considering. Starting with an option that allows for much more data storage and computing such as many of the more general cloud computing options might be best for you in this case, as you might find yourself restricted by smaller cloud platforms or more traditional shared computing resources.
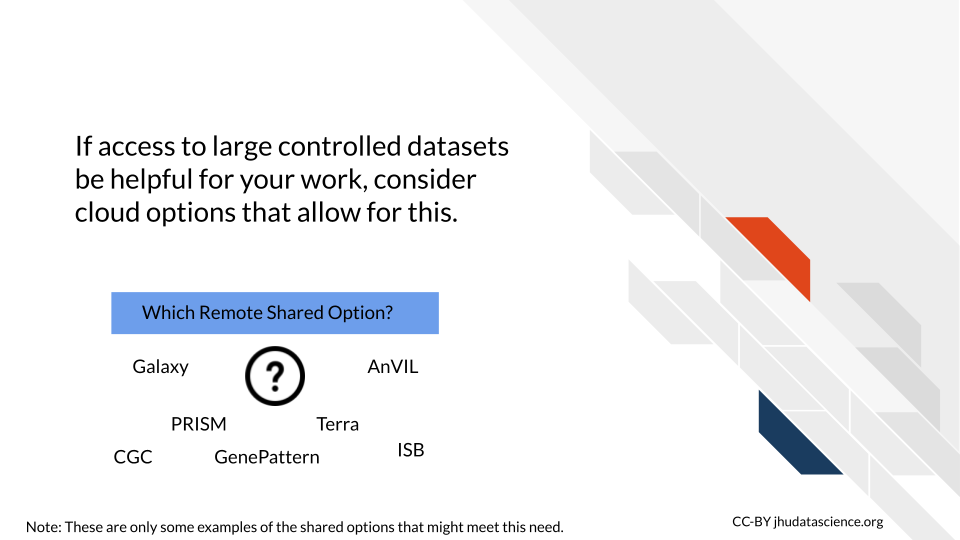
7.1.9 Data access
Some cloud computing options already have data available that may be of interest for you and your work. For example Galaxy, AnVIL, and GenePattern provide access to many genomic datasets. Smaller platforms can also have access to data that may be of specific clinical interest to you, such as the Cancer Genome Collaboratory, which provides access to data from the International Cancer Genome Consortium (ICGC).
7.1.10 Interface
Will you need a graphical interface, a command line interface, or both?
We described this a bit earlier, but here we will provide some more thorough examples.
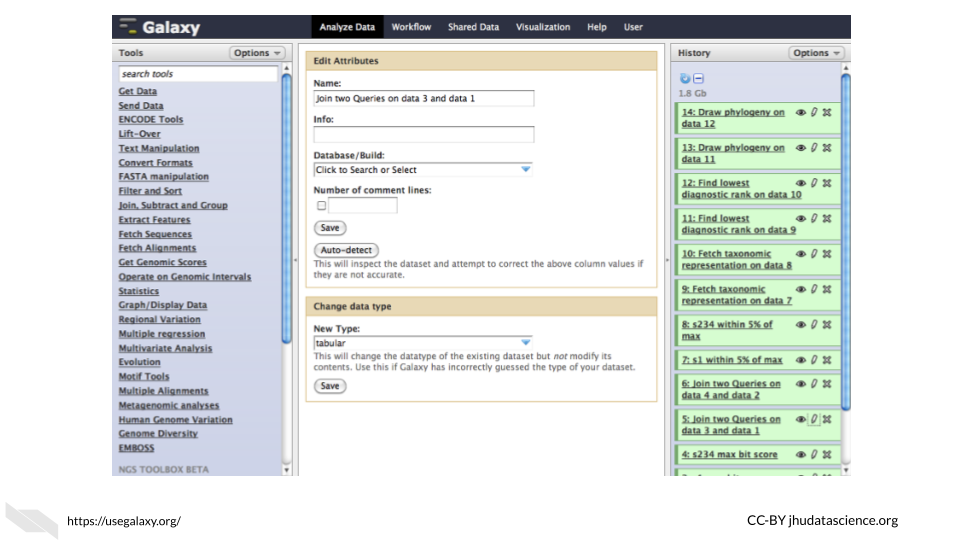
Galaxy offers a graphical user interface for performing analyses and tasks. For example in the following image we show a GUI for joining two files:
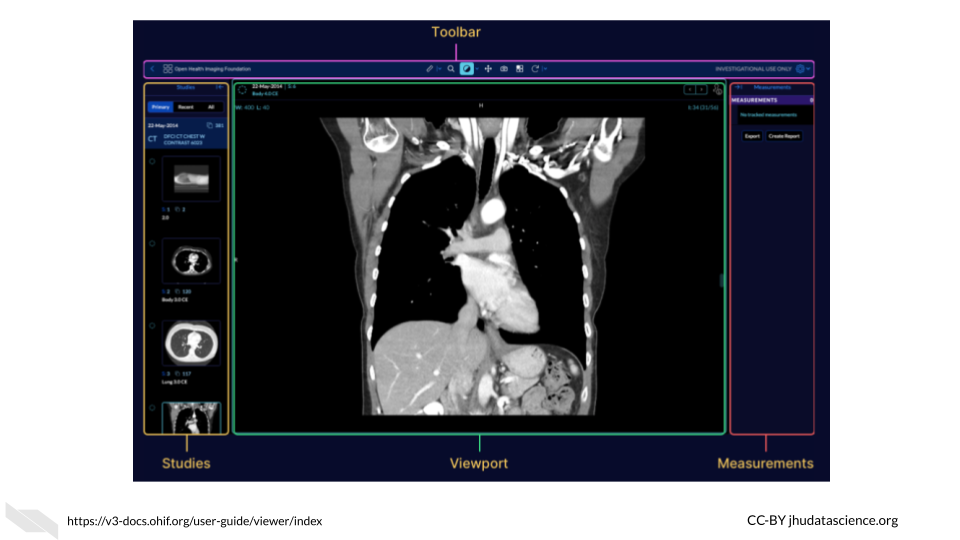
Another Graphical User Interface example comes from the OHIF image viewer which has a tool bar for modifying, viewing and annotating images.
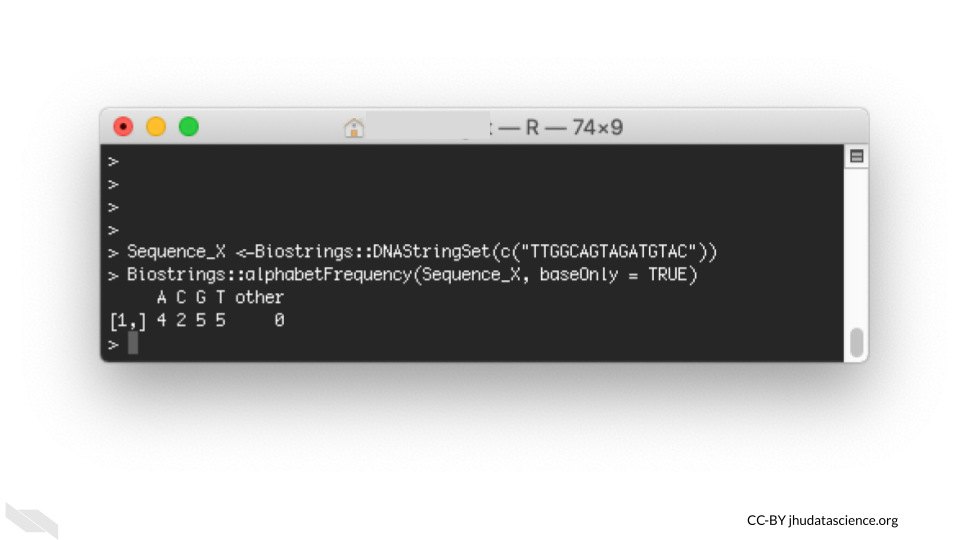
Recall that a command line interface (also known as a character interface) allows users to specify functions with code.
For example, one could perform functions in R using Bioconductor packages such as Biostrings with a command line interface:
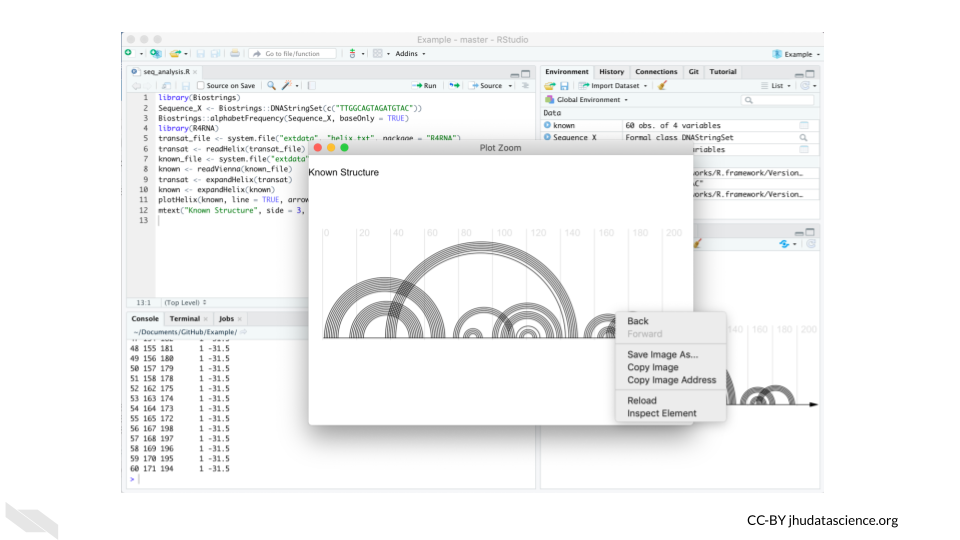
A situation where you might use both a command line interface and a GUI, is using RStudio to perform an analysis in R with Bioconductor packages. RStudio is what is called an IDE or an integrated development environment, which is an application that supports writing code. There are many tools to help you including a console for writing code in R with command line interfacing, as well as graphical interface tools. As shown in this example below, one can inspect and save a plot (that was created with the command line) by using a GUI. The plot was created using the same package as the one used in the command line interface example.
Some cloud computing options will have both interface options, while others will only have one. This is an important consideration when you decide what computing resources to use.
7.3 Choosing between remote sharing options
Should you decide that you want to go with a remote sharing option, the final major decision is to decide which remote computing resource to go with.

This decision should be based on the following:

7.4 Overall Decision Process
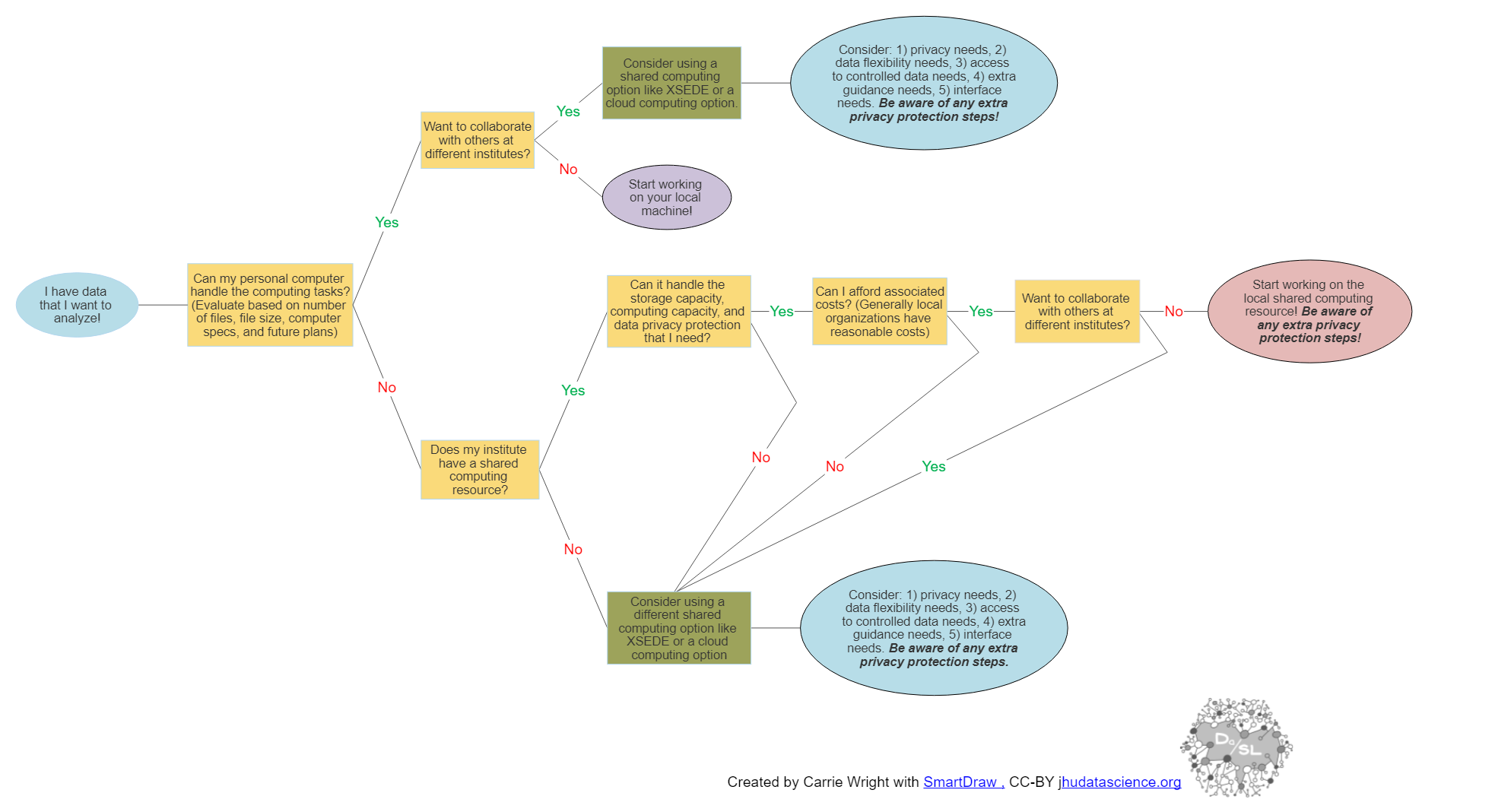
We suggest evaluating your computing needs based on the following decision tree. The tree reads from left to right, and you can click on the image to zoom.
7.5 Conclusion
We hope that this chapter has given you some more perspective on how to make important computing decisions to make the most out of your work.
In conclusion, here are some of the major take-home messages:
- The three major computing decisions are:
- Personal computer vs Shared resource
- Local shared resource vs. Remote Shared resource
- Which shared resource.
Start first with determining if your personal computer can handle your work or if you have plans to collaborate with others at different institutes.
- The following are general considerations that can help you make each of the 3 decisions:
- How computationally intensive will the work be?
- How much data storage is and will be needed?
- Do I plan on collaborating with others outside my institute?
- Does my data that need extra privacy protection?
- How much money can I spend on computing? - Do I want extra guidance for my informatics work?
- Do I need flexibility? Might I work with more data modalities in the future?
- Do I need scalability? Will I soon work with more data?
- Do I need access to large controlled datasets?
- What kind of interface would be helpful?
The main benefits of cloud options are easier sharing of data, code, and workflows even across different computing platforms, access to large amounts of computing resources, generally only need to pay for the resource you actually use, less need to be as conscious of proper etiquette for sharing computing resources.
The main drawbacks of cloud options in general are:
- If you were already using a shared resource, then migrating to the cloud will require data transfer effort and time
- Some cloud computing options do not provide data privacy protection that may be needed for certain types of data
- Costs calculations can be especially confusing and you will need to learn how it works for the particular resource that you are interested in using
- There will be less IT support form your local IT department as many of these resources have their own infrastructure, however many options provide their own guidance and support
- More and more cloud computing options are being developed and designed specifically for research, consider the following questions when choosing between cloud-based options:
- Does the resource offer the privacy protection needed?
- Does the resource allow for me to use the data modalities I need?
- Does the resource already have data that I might want?
- Does the resource provide extra guidance that might be helpful for my work?
- Does the resource offer the interface that I would like to work with?
- Does the resource offer me access to a relevant community for my work that would be helpful?