
8.4 Exercise Two: Alignment
Given that our data has passed some quality checks, we will try to align the data to the reference genome. In this case it is simple, a viral genome. A human sequencing project will generate much larger data sets. There are many aligners, but we will start off looking at a simple aligner BWA-MEM. This example uses paired data.
We will use our two SARs data files, which are ready for alignment. VA_sample_forward_reads.fastq VA_sample_reverse_reads.fastq
Now go to GENOMICS ANALYSIS: Mapping and select “Map with BWA-MEM”. This program will align your reads to your SARS reference genome. Some of our reads are >100 base pairs so we will use the MEM option.
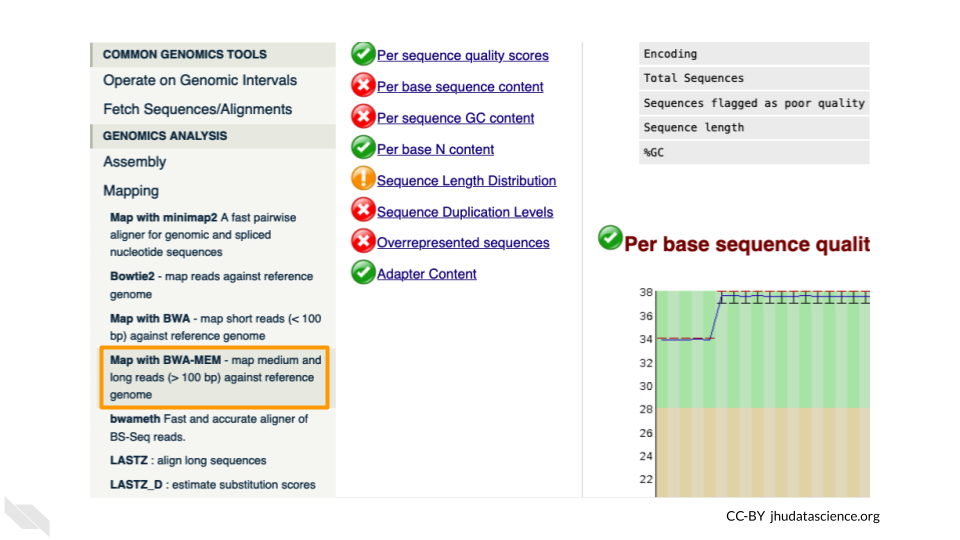
First, choose your reference. In the first drop down box change it to “Use a genome from history and build index”. Then choose the SARS reference fasta file that you uploaded as the reference.
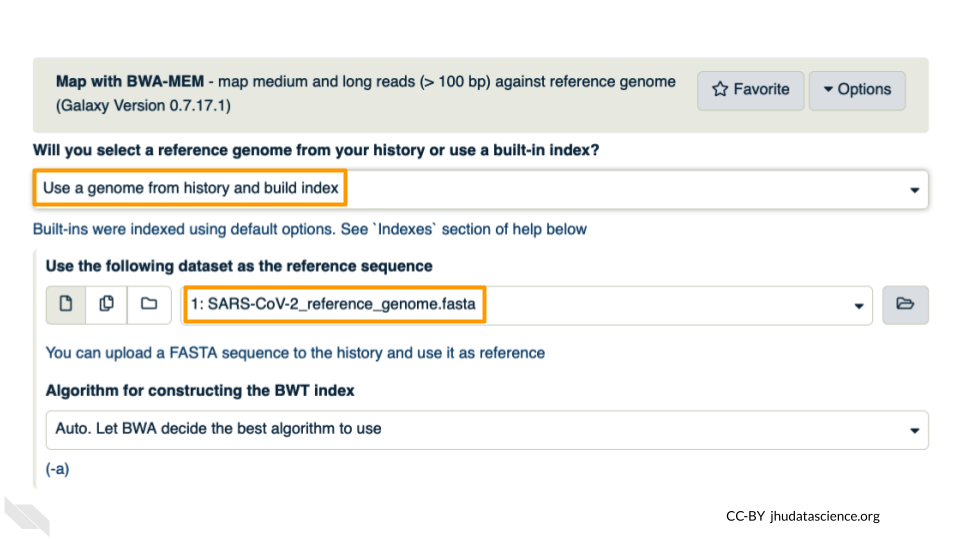
Under the “Single or Paired-end reads” ensure the “Paired” option is selected. Now choose your forward and reverse fastq files. Leave other options as-is. You can learn more about what the alignment software BWA-MEM is doing if you scroll down below the execute button. Click execute.
The output file is a BAM file, which lists where each read aligns to the reference genome and whether there are any differences. You can click the eye button to preview the results, but the results are not easy to interpret visually (much like the fastq files). Instead you will use a genome viewer in the next step.
QUESTIONS:
What is alignment software (for example, BWA-MEM) actually doing?
Here we are using paired fastq (“paired end”) data. What is an advantage of using paired data?