
8.3 Exercise One: Importing Data into Galaxy
Luckily, we linked to the original data when we cloned our Workspace! We have three files we will need for our activity. These are (1) the reference genome for SARS-CoV-2, and both forward (2) and reverse (3) reads for our sample. There are two sets of reads for our sample because the scientists who collected it used paired-end sequencing. The reference genome ends in “.fasta” because it has already been cleaned up by scientists. The sample we are looking at ends in fastq because it is raw data from the sequencer.
Click on “Upload Data” in the Tools pane.
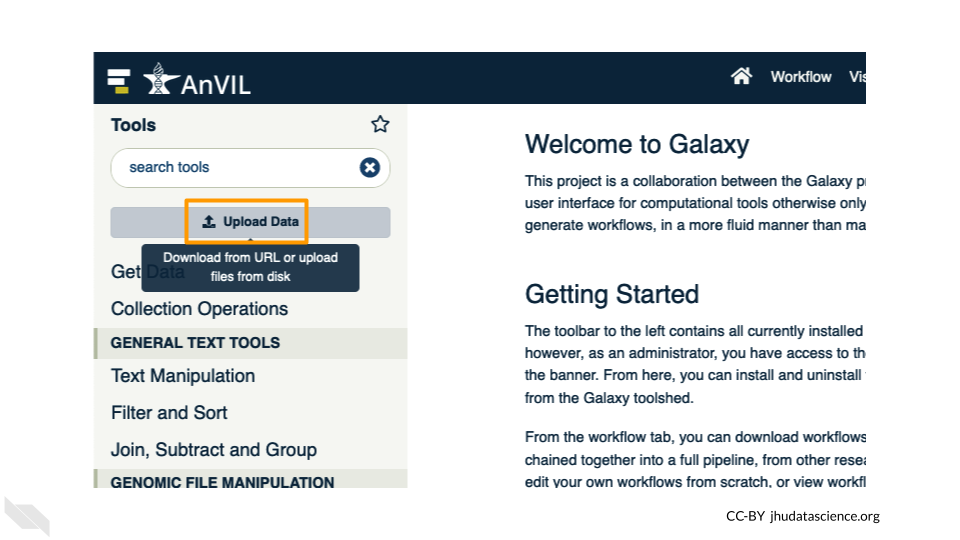
Click on “Choose remote files” at the bottom of the popup. Double-click the workspace folder, then “Tables/” then “reference/”. Click the reference .fasta file so that it is highlighted in green and click “OK”.
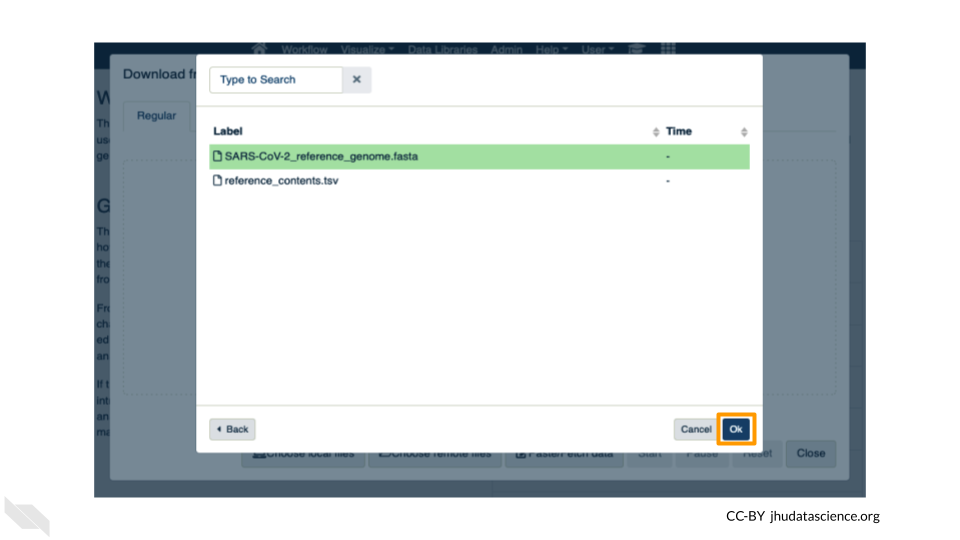
Now that your reference has been added, click “Choose remote files” again to add the two sample files. Double-click the workspace folder, then “Tables/” then “samples/”. Click the two sample
fastqfiles so that they are highlighted in green and click “OK”.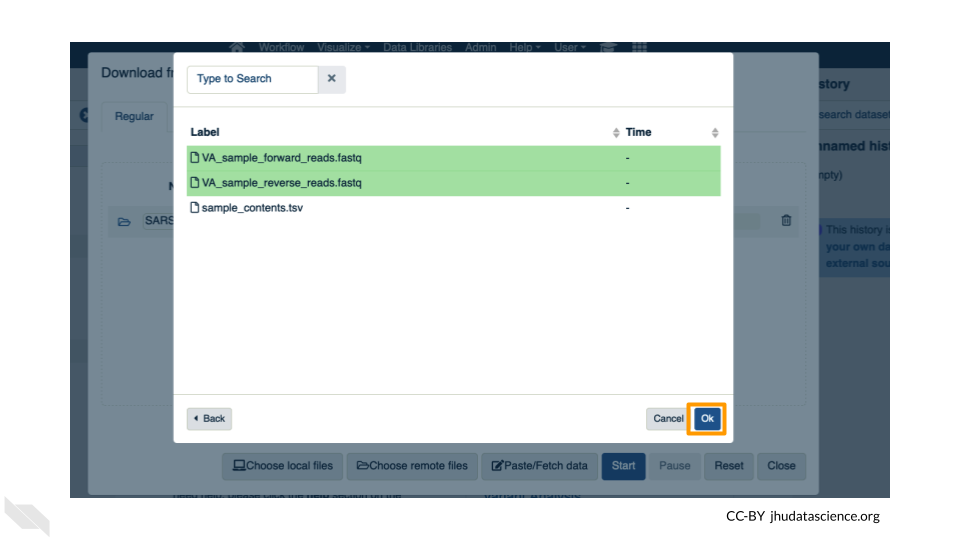
Click “Start” and once complete, you can click “Close”.
Confirm your upload worked by looking at the file names in the History pane.
